Top Links
Journal of Veterinary Science and Animal Husbandry
ISSN: 2348-9790
Salmonella Serovars and Their Host Specificity
Copyright: © 2013 Vikash Singh. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Related article at Pubmed, Google Scholar
Salmonella is a causative agent for a wide variety of pathological diseases in humans, cattle, poultry and other farm animals and hence Salmonella infections are a major cause of concern to humans, veterinary animals and to food industry. With characterization of over 2500 Salmonella serovars, the pathogen nearly infects all vertebrates but the severity of infection varies from one serovar to another depending upon their host specificity. Some Salmonella serovars are restricted to one or few hosts while others have a broad host spectrum. Thus the understanding of the mechanisms involving host preference by one serovar over another is very important. As our knowledge about host adaptability will then be instrumental in designing better vaccines. Furthermore, methods involving identification of genetic markers for host specificity will prove to be instrumental in determining virulence factors for other pathogenic bacteria that cause systemic infections.
Keywords: Host adaption; Evolution; Salmonella serovar; Horizontal gene transfer
Salmonella species belong to Gram negative, rod shaped, facultative intracellular bacteria that successfully infects a wide variety of hosts. Salmonella not only infects humans, but also numerous farm animals; including pigs, cattle, horses and chickens. Hence, the infection caused by Salmonella poses a serious threat, not only to farmers but also to the food industries, especially in countries where meat is the most preferred food. Salmonella is considered to be a 'universal pathogen' as it is successfully isolated from all vertebrates and many insects [1].
Salmonella infections vary from asymptomatic colonization in the gut to inflammation, diarrhea or typhoid fever [2], depending upon the serovar and its host preference. 'Serovar-host adaptation' or 'serovar-host specificity' refers to the prevailing differences between the Salmonella serovars with respect to their host preference and clinical manifestations [3,4]. These preferences depend upon the ability of each serovar to adapt itself to the environment within its host [5]. These adaptations are attributed to numerous virulence determinants and other microbial characteristics of a particular serovar which makes it specific for its host. Recent studies show that a mechanism making one serovar virulent for one animal species could make the same serovar less or completely avirulent in another animal host [6]. In addition, other factors like the dose of infection, the age during which the host is infected and their immune response contribute equally to a successful infection [7]. Even though the complete genome sequences of several Salmonella serovars are available, the molecular mechanisms defining Salmonella colonization, its host preferences over the other, its pathogenesis and transmission of the infection to other hosts still remains unclear.
Here, in this review we focus our understanding on various factors and mechanisms acquired by different Salmonella serovars for host specificity and adaptation.
Horizontal gene transfer has played an instrumental role in the divergence of Salmonella from its ancestors. Molecular methods used to study epidemiology among Salmonella serovars suggests that Salmonella spp has diverged from E.coli lineage [6,8,9] and this evolution could be characterized into 3 distinct phases
The first phase epitomizes the acquisition of SPI-1 (Salmonella Pathogenecity Island-1) pathogenic determinant through horizontal gene transfer. This pathogenic determinant codes for genes and virulence factors responsible for the invasion of the Salmonella within the host, by bringing about re-modulation of the host cytoskeleton [10]. Interestingly, SPI-1 is present in all the Salmonella serovars and is absent in E.coli and other related gram negative bacteria.
The second phase marks the emergence of two distinct Salmonella species, Salmonella enterica and Salmonella bongori, as a result of horizontal gene transfer. The species S. enterica acquired the SPI-2 pathogenic determinant (island), which is responsible in establishing a niche for the bacterium within the host, thus helping the Salmonella in surviving intracellularly after successful invasion in a wide variety of hosts including the macrophages [11].
The third phase is seen as the division of the Salmonella spp. into sub species and their adaptation to their respective hosts. The Salmonella enterica spp. is further divided into six sub species namely: S enterica subspecies enteric, S enterica subspecies salamae, S enterica subsp. Diarizone, S enterica subsp. Houtenaae, and S. enterica subsp. Indica [8]. The S enterica subspecies enterica got adapted to warm blooded vertebrates (birds and mammals), while the sub-species of Salmonella bongori are mainly adapted to cold blooded vertebrates.
Thus, understanding of the biological phenomena and molecular mechanism leading to the development and survival of these serovars within their hosts is of high importance. Hence, current research is mainly focused on understanding the acquired ability of Salmonella's host preference by Salmonella.
Based on the surface 'O' antigen, which is a part, of the variable long chain of lipopolysaccharide on the bacterial outer membrane, more than 2,500 different serovars of Salmonella has been characterized [10]. Out of these 2,500 serovars nearly 1500 belong to the Salmonella subsp. enterica. Figure 1 represents different Salmonella serovars with core genome and with unique genes marked in black [12].
Serovars of the enterica sub species can be divided into three groups (Table 1) depending upon their ability to infect a wide variety of hosts. The first group includes serovars which have a broad host range also called as unrestricted serovars as these infect nearly all animals. This group includes serovars like Salmonella Typhimurium and Salmonella Enteritidis. These serovars cause mild enteric diseases, or usually, these persist within the host without any severe clinical symptoms [13]. Although the severity of disease increases in young hosts when compared to adults, this is because of their inability to counter the mature immune responses in older hosts [8]. Nevertheless, these serovars are of high importance with respect to their epidemiology as these have developed mechanisms to invade different hosts without any greater resistance. Thus, these serovars pose a greater zoonotic potential than their other counterparts.
The second group includes serovars which cause highly severe systemic infection in their preferred host and are usually excreted without any clinical symptoms when they accidentally infect hosts others then their most adapted or preferred. Serovars such as Dublin, Choleraesuis fall into this category, as these prove to only cause systemic infection in cattle and pigs respectively [13,14]; however these upon infection into other hosts like rodents and humans are usually excreted making these hosts as 'carriers'. Serovars of this group are referred to as the 'Host-adapted Serovars'.
The third group comprises of serovars which are restricted very strictly with one very specific host only; these serovars are called 'host–restricted serovars'. They exclusively cause systemic infection, which often proves to be fatal within their host. Serovars such as Typhi, Gallinarum, Abortusequi etc belong to this group. These serovars modulate their host's natural environment according to their benefit by exhibiting high trophism for lymphatic organs of their host; [15] Salmonella Typhi is a classic example that is associated with this phenomena. Another striking feature of serovars in this group is their ability to proliferate in fetal tissues, thus affecting egg production in poultry and causing abortions in mammals [15].
Salmonella infections are basically contracted due to ingestion of contaminated food and water. Upon ingestion, the bacterium multiplies in the small intestine and within a few weeks, it invades the intestinal wall and spreads to other organs in the body [16, 17]. Salmonella thrives on the Payer's patches, which is abundant with specialized epithelial M cells, and are considered as the primary site for infection. Upon breaching the mucosal layer, it then translocate to lymphoidal follicles and mesenteric lymph nodes [17].
Salmonella has developed mechanisms to infect and proliferate both in phagocytic and non-phagocytic cells. These include the epithelial cells, macrophages, dendritic cells, enterocytes and neutrophils [18]. The entry of Salmonella within cells is either by phagocytosis, Salmonella mediated through Type Three Secretion System-1 (T3SS1) or T3SS1 independent [10]. The process involves secretion of virulence factors called effector proteins encoded by SPI-1, which bring about actin re-modulation, leading to ruffling and extension of the plasma membrane of the host and hence resulting in invasion of the bacterium [10, 19, 20]. Once inside the epithelial cells, Salmonella develops around it a niche called the Salmonella Containing Vacuole (SCV). These SCVs interact with the endocytic vesicles within the host, thereby accumulating various factors in the process [21]. These include Rho GTPase such as Rab5 and Rab7 and also lysosomal associated membrane protein LAMP-1 [22]. From the SCV, the bacterium secrets another set of effector proteins encoded by SPI-2 genes that are responsible for intracellular replication and survival [23,24]. After 4-6hrs of invasion the replicating bacteria within the SCV results in formation of tubular network like filaments called the Salmonella induced filaments (Sifs), which helps in maintain the integrity of the SCV [23]. These Sif's tend to grow outwards [21] the plasma membrane accumulating various host constituents. The formation of these Sif's is facilitated by TTSS-2 effector protein called SifA [21, 23, 24]. These Sif's are highly enriched in cholesterol and LAMP-1. Internalization of Salmonella, also affects other cellular process such as apoptosis, cell division, cytokine production and antigen presentation [25].
Successful adaptation to a host by any pathogen is a complex process as it involves numerous factors and genetic determinants which are a result of large number of gene products. Although the precise mechanisms leading to host specificity by Salmonella is not very well understood, however the pathogenicity of Salmonella serovars is influenced by selective pressure within a particular host and its surroundings [5,8].
The most common strategy that most pathogens adopt in order to establish themselves within their host involves either acquisition of novel virulence determinants and plasmids or they tend to reduce their genome. Serovars such as a Salmonella Typhimurium, Enteritidis, Pullorum, Gallinarium Dublin and Paratyphi C are a classic example which has undergone gene deletions [13]. As a result, these serovars have lost the ability to replicate in the intestinal lumen of their respective host, although these successfully cause systemic infections [26]. In addition to, gene deletion, acquisition of various phage elements containing virulence determinants through horizontal gene transfer also plays a significant role in host adaptability by Salmonella serovars. Attachment to host cell surface is a pre-requisite parameter to be considered for a successful invasion of Salmonella with in its host. Mannose sensitive pathogenicity determinants like FimH adhesins play an important role in adhesion of Salmonella on its host cell surface [27]. Therefore, any allele variations or point mutations resulting in structural modification of these adhesions would be an important determining factor leading to host-specificity [27]. For example, type I FimH adhesions are found to be expressed by Unrestricted serovars which infect both mammals as well as chickens, however, type-2 FimH adhesions are expressed only by Host-Restricted serovars such as Salmonella Gallinarium [27].
Apart from genetic factors, other paradigms such as physiological state of host cell, availability of amino acids and the ability of one serovar over other to replicate, has a critical role to play in the virulence pattern of a serovar [17]. For example, S. Typhimurium showed rapid and better replication in swine ileal mucosa over S. Choleraesius. As a result of its better replication the S. Typhimurium serovar generated significant amount of immune responses leading to its faster clearance whereas the slow growing S. Choleraesius was successfully able to disseminate the immune response, eventually leading to systemic infection in swine [19].
Regardless of various genetic and physiological parameters effecting host specificity, it is also observed that stress has a significant role to play in pathogenicity and virulence of Salmonella in various hosts leading to its consistent presence in the food chain and environment. One of the classic examples is increased fecal shedding of S. Typhimurium in pigs, cattle and other farm animals upon transportation [28, 29]. This form of social stress in the host leads to reactivation of asymptomatic infection and is often considered to have a high zoonotic potential. Such phenomena of reemergence could be partially explained by Salmonella's ability to respond to host stress as during transportation the farm animals secrete stress hormones like Noradrenaline which eventually results in excretion of iron from host storage compartments and proteins which is used by Salmonella for its growth and hence the fecal shedding [14,30].
Apparently, it appears that in depth study is needed taking in to account both host as well as pathogen determinants in order to have a clear view about the numerous factors and parameters governing host-specificity of a particular serovar.
Despite our understanding of the molecular mechanisms of Salmonella pathogenesis, the factors and fundamental mechanisms required by a serovar to exhibit host specificity and preference for a particular host remains unclear. However, recent studies involving immune responses upon infecting two serovars in the same host, suggests that the host adapted serovar competitively excluded the other serovars in a given population. However, assessing the zoonotic and epidemiological characteristics of different Salmonella serovars will be instrumental in estimating the degree of risk posed to humans.
Improvements in sequencing methods have shown insights in specifically highlighting clinical manifestations associated with one particular serovar but absent in the other. Methods like SNP typing and MLST have paved the way for faster and precise serotyping of different isolates of Salmonella from various farm animals. Our in-depth understanding of host specifity of one serovar over the other would pave the path in designing better vaccines which would eventually prove to be effective against the most dominant serovar in the farm animals. This would hopefully help in the improvement of the efficacy of the vaccination process. Therefore, further research should focus upon the identification of genetic markers for the origin of host adaptation and host specificity. Such studies would help the researcher to predict a serovar's survival in a new animal population, thus enabling them with tools to design better strategies against the most prevalent serovar in the livestock. Furthermore, these methods could then be used as a model in determining virulence factors used by other pathogenic bacteria in causing diseases to specific animal hosts.
| Group | Serovars | Host | Infections |
| Unrestricted Serovars | S. Typhimurium S. Enteritidis |
Humans, Poultry Cattle, Swine Mouse |
Enterocolitis in humans and Swine Asymtomatic carriers in Poultry and Cattle Septicemia in mouse |
| Host Adapted | S. Dublin, S. Cholerasuis |
Cattle, Pigs , rarely in Humans, Mouse and Chickens |
Septicemia, Enterocolitis in Cattle Fatal systemic infection in swine Bacteremia in Humans and Mouse. |
| Host restricted | S. Typhi S. Gallinarum S. Abortusequi |
Humans Poultry Horses |
Typhus, Diarrhea Septicemia, fatal Leads to abortions in mares |
*-- not detectable
Table 1: Classification of Salmonella serovars as per their host specificity and type of infection caused by them in their host.
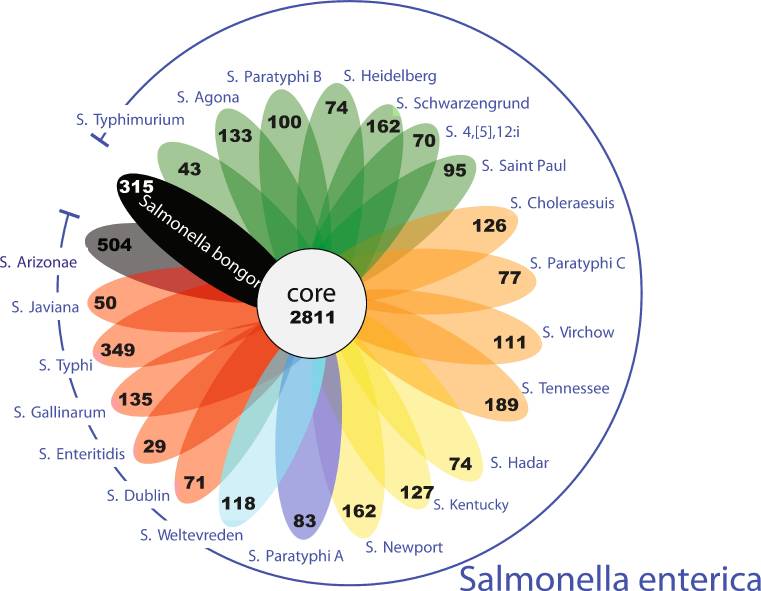 |
| Figure 1: A flowerplot representing unique gene families in each Salmonella serovar with core genome marked in the centre (adapted from [12]). |






































