Top Links
Journal of Proteomics and Genomics
ISSN: 2576-7690
Dynamics of Protein Tyrosine Nitration and Denitration: A Review
Copyright: © 2025 Sengupta S. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Related article at Pubmed, Google Scholar
Protein tyrosine nitration (PTN) is a post-translational modification that occurs under the action of Nitric oxide (NO) which leads to the formation of reactive nitrogen species (RNS). RNS reacts with tyrosine residues in proteins to form nitrotyrosine. PTN has lately been related to neurodegenerative diseases. It is very essential to know the details of its mechanism. PTN is also involved in cellular signaling. The detail of such an event is still not clear. Controlling of cellular signaling is only possible if nitration and denitraton is a dynamic, regulated and reversible process. In this review the biological consequence of PTN, analytical methods required for their detection and their relation with different neurological diseases has been discussed in details. This review is the first of its kind where the use of bioinformatics tools for detection of nitrotyrosine has been discussed comprehensively. The review gives a detailed account of the existing softwares, databases and their uses for the study of PTN and denitration. Moreover the functioning of the probable denitration pathways and associated impacts on cellular signaling and physiology have also been addressed in detail. Although previous reviews on PTN are limited only to its formation, function, analysis or denitration of nitrated proteins separately but integrating them to provide the broader picture and relevance in biological system was absent. This work is aimed at to bridge that lacunae also provide insights about the identification and detection of interactive components in a protein nitration-denitration pathway.
Keywords: MProtein Tyrosine Nitration; Nitric Oxide; Neurodegenerative Diseases; Denitration; Bioinformatics Tools
List of Abbreviations: NO2-TYR: Nitrotyrosine; RNS: Reactive nitrogen species; NO: Nitric oxide; PTN: Protein Tyrosine Nitration; GSH: Reduced glutathione; GSSG: Oxidized glutathione; MnSOD: Manganese superoxide dismutase; HDAC2: Histone Deacetylase 2; COX: Cyclooxygenase; LDL: Low-density lipoprotein; HDL: High-density lipoprotein; HPLC: High Performance Liquid Chromatography; NH2-TYR: Aminotyrosine; GC–MS: Gas Chromatography-Mass Spectroscopy; 2DGE: Two-Dimensional Polyacrylamide Gel Electrophoresis; LC-MS/MS: Liquid chromatography-tandem mass spectroscopy
Stress is a situation in which the cellular redox homoeostasis is altered because of excessive production of different reactive species eg. reactive oxygen species (ROS), reactive nitrogen species (RNS) [1]. The stress which is mediated by ROS like singlet oxygen, superoxide, H2O2 and hydroxyl radicals known as oxidative stress [2]. ROS are produced during cell cycle progression, cell differentiation, cell signaling [3,4]. It can damage the macromolecules like membrane lipids [5], proteins [6] and DNA [7]. In analogy to oxidative stress, the term `nitrosative stress' involves reactive nitrogen species (RNS) where the ratio of nitrosants to antioxidants is >1. RNS are produced due to the reaction of ROS with Nitric oxide (NO•). During nitrosative stress different types of RNS cause nitrosylation of biomolecules thereby creating an imbalance in the production and the exclusion of reactive nitrogen and oxygen intermediates of the body [8]. One of the major marker of nitrosative stress is the formation of 3-Nitrotyrosine (NO2-TYR), a stable post-translational modification of protein. In vivo it forms due to the reaction between tyrosine residues and nitrating agents. During the formation of 3-Nitotyrosine a nitro group (-NO2) is added in the ortho position of the phenolic hydroxyl group of tyrosine. Mostly the natural abundance of tyrosine residues is about 3% in proteins. Tyrosine may be nitrated through numerous chemical reactions. This modification introduces a net negative charge at neutral pH, which triggers changes in the local physiological and chemical environment of the biomolecule. Nitration of tyrosine residues often indicates loss of protein activity. Alteration of the structure and function of protein due to formation of nitrated tyrosine may change the rate of proteolytic degradation [9]. Moreover presence of high amount of RNS or non-functional antioxidant systems, target proteins can also be nitrated at specific tyrosine residues [10].
3-Nitrotyrosine formation is associated with cell signaling and disease initiation and progression like neurodegenerative diseases, cardiovascular injury and cancer. It even facilitates in the accumulation of nitrated proteins in vivo [11]. The most common neurodegenerative diseases are Alzheimer's disease, Huntington's disease and Parkinson's disease etc. These Diseases are also referred to as proteinopathies due to the aggregation of proteins [12]. Loss of synapses and neurons in the certain subcortical regions and cerebral cortex is a characteristic feature of Alzheimer’s disease. This loss indicates the degeneration of parietal and temporal lobe of the brain and also parts of the cingulate gyrus and frontal cortex [13]. Alzheimer's disease is triggered by accumulation of misfolded αβ and nitrated tau proteins in the brain [14]. Accumulation of intracellular toxic proteins and late onset are both essential features of Huntington’s disease and Parkinson’s disease. Not only PTN is related to diseases but it is also related to cellular signaling. Different proteins of signaling pathway of mating type Saccharomyces cerevisiae is nitrated [15]. Beside these the mitochondrial proteins like aconitase, isocitrate dehydrogenase are also reported to be nitrated in Saccharomyces cerevisiae [16]. Thus the accumulation of only nitrated proteins subsets is dependent on the conversion of nitrated proteins to its denitrated form and vice-versa [17]. But the evidences of denitration process are very less. The fine tune between the balance of nitration and denitration is yet to be elucidated. Although PTN as of now is a stable post-translational modification, but the role of nitrated protein in cellular signaling and physiology is slowly emerging.
Several biochemical systems are present to counteract the nitrosative stress [1]. Among them Low molecular weight thiols play an important role to counteract the nitrosative stress and also maintain the redox homeostasis. One of the important low molecular weight thiol is reduced glutathione (GSH) which is oxidized to GSSG by the action of glutathione peroxidase (GPx) in the elevated amount of NO• to maintain the cellular homeostasis. GSSG is again converted back to GSH by the activity of glutathione reductase. The ratio of the concentration of GSH and GSSG is associated with the redox homeostasis [18]. Macromolecules like enzymes can also counteract stress condition [1]. Catalases can counteract the activity of RNS. It is reported that catalases can detoxify the effect of peroxynitrite in vivo [19]. Superoxide dismutase [20], Cytochorme C [21] are also reported as the antinitrosative enzymes but the activity of these enzymes are also inhibited by nitration [22,23]. Thus the antinitrosative and denitration system is a topic of ongoing research.
In this review we have discussed about the cellular function of PTN and relation of PTN to the neurodegenerative diseases and we have attempted to give a hypothesis to arrest the denitration system (i.e. the system by which organisms can counteract the PTN) by using the proteomics tools [15,17].
NO• is an endogenously and enzymatically generated molecule which contributes diversely to physiological functions [24]. NO• is a very reactive molecule as it contains unpaired electron that permits rapid reaction with diverse molecules [25] and form reactive nitrogen species that can nitrify the protein tyrosine [26]. NO• is formed during the Conversion of L-arginine to L-citrulline. The first step is the formation of intermediate Nω-hydroxy-L-arginine (NHA). This reaction is catalyzed by Nitric oxide synthases (NOSs). NHA is again oxidized to form NO• [27]. The post-translational modification through NO• is a non-classical NO• signaling, which is cGMP-independent [28]. But NO. also plays an important role as signaling molecule [29] in neurotransmission, vasodilation [30], and immune response in vivo therefore regulation of the activity of NOSs (specially iNOS) to synthesis NO•, is associated with many diseases [31-35].
Peroxynitrite (ONOO-) is synthesized quickly on reaction of nitric oxide (NO•) with superoxide anion (O2-•) [36]. Superoxide is produced in various biochemical reactions because of the electron flow through the mitochondria which can interact with NO to form peroxynitrtite [37]. Peroxynitrite can also be synthesized in vivo through the reaction of molecular oxygen with nitroxyl anion (NO-) [38]. Most of the time peroxynitrite reacts with carbon dioxide in cellular fluids and synthesize ONOOCO2-, which is decomposed by two ways- one gives carbon dioxide and nitrate, and the other carbonate radical anion and nitrogen dioxide (NO2). The second way is the key pathway of protein tyrosine nitration via peroxynitrite. This adduct of peroxynitrite-carbon dioxide may cause nitration directly via NO2+ [39] (Figure 1).
Nitrates and nitrite can be another source of nitric oxide apart from L-Arg nitric oxide synthases. There are several enzymatic as well nonenzymatic pathway to from NO• from Nitrate and nitrite in vivo [40]. Nonenzymatic NO• formation from nitrate esters involves interaction with thiol groups. Further, thiols are oxidized to their respective disulfides and nitrite. A sigmoidal thiol dependence was found for NO• formation, suggesting a more complex reaction mechanism. The rate of reaction between nitrate and thiol increases at alkaline pH [41]. Protein tyrosine can be nitrated by NO2• radicals. This radical is formed through the reaction of nitrite and hydrogen peroxide by various hemoperoxydases. It is one of the key mechanism of PTN [42]. Nitrous acid is formed by acidification of nitrite and sometimes nitryl chloride (NO2Cl) [43]. This nitryl chloride is a potent nitrating agent. Certain metalloproteins comprising hemoglobin, cytochrome c, superoxide dismutase catalyze H2O2 dependent oxidation of nitrite to form 3-nitrotyrosine [44].
Exposure of GSH to •NO can lead to formation of S-nitrosoglutathione (GSNO) [45]. GSNO is a source of •NO, and its plays an important role in •NO mediated signaling. S-nitrosothiol can also imitate the effects of endogenous •NO because •NO is released during GSNO decomposition [46]. It has already been reported that GSNO can up regulate the activities of catalase and SOD (Superoxide Dismutase) to combat oxidative/ nitrosative stress in yeast [47]. In vivo S-nitrosothiols has an important role in controlling protein function and structure together with also in •NO transport and signaling [48]. Redox homeostasis is maintained by glutathione (GSH). In case of mammalian cells glutathione is involved in transport of amino acid, detoxification of xenobiotic and deoxyribonucleotides and inhibition of platelet activation [49-51].
Protein tyrosine nitration may alter the function of proteins [52]. Tyrosine though may be the major target for nitration but cysteine and methionine can be nitrated too [53]. For example in the case of MnSOD (Manganese superoxide dismutase), substrate (O2•−) is diffused towards the Mn center due to the steric restriction which is imposed by −NO2 group. It causes a fall in the pKa of the –OH group of tyrosine. In the active site a negative charge is formed due to the nitration of a tyrosine residue, which results in the electrostatic repulsion of O2•− and alteration of the redox potential and arrangement of hydrogen bonding [54]. It was reported that the peroxynitrite can inhibit the activity of MnSOD of Arabidopsis by nitration at Tyr63 residue [22]. In the nitrating conditions cytochrome c results in nitration at adaptable positions cytochrome c tyrosine. Whereas nitration of cytochrome C by peroxynitrite causes preferential modification of tyrosine residue, leading to mono nitrated forms of tyrosines moderately distant from the active site [54]. Reports suggest that the activity of HDAC2 (Histone Deacetylase 2) is inhibited due to the tyrosine-253 nitration. This nitration also causes prompt proteasomal degradation of HDAC and due to this event gene regulation is affected in vivo. This study explores HDAC inhibitors as the novel cancer therapies [55]. In a recent study NF-κB mediated mechanism has been proposed where nitration of tyrosine-181 of IκBα results in activation of nitric oxide synthase constitutively, which causes dissociation of intact IκBα from NF-κB and this mechanism doesn’t seem to involve proteolytic degradation of IκBα or IκBα kinase-dependent phosphorylation [56]. This activation of NF-κB leads to inflammation [57]. In another study activation of carbamoyl phosphate synthetase 1(CPS1) is prevented by activator N-acetyl-L-glutamate (NAG) due to the nitration at Y1450 in an α-helix of allosteric domain [58]. One of the major enzyme Ascorbate peroxidase regulates ascorbate–glutathione cycle which is very important for the detoxification of cellular H2O2. But the activity of Pea Ascorbate peroxidase is inhibited due to the protein tyrosine nitration. It is revealed that Tyr5 and Tyr235 residues were nitrated by peroxynitrite [59]. A study revealed that high amount of NO. can inactivate NADPH oxidase thus self-amplification loop is slowed down [22].
Specific tyrosine residues of diverse proteins are nitrated both in vivo and in vitro condition by the reactive molecules like peroxynitrite [60]. The presence of 3-nitrotyrosinated proteins is apparent histochemically in infected or inflamed tissues. Protein nitration may be profoundly associated to oxidative cell injury indicating its role in altering the protein function. The structure of the proteins is also changed due to the nitration and the distorted proteins may act as antigens thus antibodies are generated against self-proteins [61]. The immune reactivity of 3-nitrotyrosine has been conveyed in numerous human pathological condition. Increased levels of 3-nitrotyrosine is associated with different human diseases such as multiple sclerosis, atherosclerosis, Parkinson’s disease (PD), Alzheimer’s disease (AD), amyotrophic lateral sclerosis (ALS), stroke, cystic fibrosis, lung diseases, asthma, myocardial malfunction, diabetes, cirrhosis, chronic hepatitis, etc. [62].
Alzheimer’s disease: Nitration of the tau protein is the cause of Alzheimer’s disease [63]. It is characterized by neurofibrillary tangle formation due to tyrosine nitration and self-assembly of tau protein [64]. Tau is a microtubule associated protein (MAP) encoded on chromosome 17. Splicing leads to the formation of six canonical isoforms in the central nervous system containing different N-terminal inserts and microtubule binding repeats [65]. Endogenous tyrosine residues prone to be nitrated in tau protein are at 18, 29, 197, 310, and 394 position (numbers correspond to the longest tau isoform, which contains441 amino acids). In vitro, selectively of ONOO- mediated tyrosine nitration is mostly seen at N-terminal region of the tau molecule (Y18, Y29) and less at Y197 and Y394. Nitration at Y310 was rarely observed, probably because of its location within the hydrophobic microtubule binding repeat of tau [66]. Generation of ONOO- is associated with αβ plaques formation [67]. αβ accumulated in mitochondrial matrix and affect the mitochondrial enzymes. So there is a decrease in brain energy metabolism [68].
Parkinson’s disease: The main cause of PD is tyrosine nitration of α-Synuclein (α-Syn) protein. All the four tyrosine residues of α-Syn are susceptible to be nitrated which are located at 39 (at the N-terminal region), 125, 133 and 136 (at the C-terminal region) [69]. α-Syn is very sensitive to nitrating agents and the protein is nitrated even at low concentration of peroxynitrite. In the presence of peroxynitrite the tyrosine residues are oxidized to form dimers which lead to oligomer formation [70]. After exposing to nitrating agent, elevated rate of dimer and oligomer formation is mediated via Tyr-125 and Tyr-39 nitration respectively [71,72]. The rate of fibril formation is accelerated by Monomeric or dimeric nitrated α-syn but the fibril formation is inhibited by nitrated α-syn oligomers [73].
Atherosclerosis: Another major disease resulting from protein tyrosine nitration is Atherosclerosis [74]. Approximately 0.001 to 0.01% tyrosine residues of plasma proteins are found to be nitrated in cardiovascular disease [75]. In this disease COX (Cyclooxygenase) gets nitrated [76]. COX plays an important role in prostaglandin biosynthesis. The enzyme catalyses the metabolism of arachidonic acid which is a key step in prostaglandin biosynthesis. A heme dependent nitration of Tyr385, present at the catalytic site of COX results in its inactivation [77]. Such nitration may also render the protein susceptible to proteolysis [74]. Different studies showed that nitration of LDL (Low-density lipoprotein), HDL (High-density lipoprotein), can lead to plaque formation, increased platelet aggregation, clotting and disruption in cholesterol transport which may also lead to atherosclerosis [74,78,79].
Nitration of tyrosine may interfere with the formation of tyrosine phosphorylation, which may affect cellular functions [80]. In vivo signal transduction is regulated by tyrosine phosphorylation. It is also associated with different cellular responses and growth factors. The neutral –OH group of tyrosine is phosphorylated by tyrosine kinases. The pka value of the –OH group of tyrosine gets shifted due to nitration thereby affecting tyrosine phosphorylation [17]. Sometimes a reverse situation can also be seen. The phosphorylation of protein tyrosine can constrain the PTN. A study showed that the phosphorylation of tyrosine of cdc2 is prevented by peroxynitrite-mediated PTN [82]. Nitration in p130Cas, an adhesion complex protein, prevents tyrosine phosphorylation, indicating that the assembly of focal adhesion complexes may get affected by this process [83]. It is also seen in different studies that the tyrosine sites are different for nitration and phosphorylation in a same protein. Tyrosine residue in extracellular signal-related kinase (ERK) 1/2 can be nitrated by stimulation of Angiotensin II which may facilitates the activation of phosphorylatio but the sites for phosphorylation and nitration is different [84]. A very present study revealed that the phosphorylation of PLK3 at S129 cannot be prevented by the nitration of α-syn at any tyrosine residue and also PLK3 phosphorylates nitrated dimers, trimers, and oligomers [85]. Hence the general idea is that protein tyrosine nitration and phosphorylation are mutually exclusive process which plays an important role in cellular signaling [86]. Tyrosine phosphorylation is dependent on the concentration of nitrating agents like peroxynitrite. Competition occurs for the same sensitive tyrosine residue for phosphorylation or nitration. While tyrosine phosphorylation is induced at lower concentration of peroxynitrite, at relatively higher concentration the process gets inactivated and it is also irreversible. But till now the competition-correlation between phosphorylation and nitration is poorly understood [87,88] (Figure 2).
The primary locus for PTN in mitochondria is matrix proteins [89]. Different reactive oxygen species are formed during the electron transport chain in mitochondria [90] which may react with nitric oxide to form Peroxynitrite. Peroxynitrite thus formed, can diffuse from extra mitochondrial compartments into mitochondria or it may be formed intramitochondrially [91]. Aconitase, a key enzyme of TCA cycle is inhibited by peroxynitrite. It disrupts the enzyme’s Fe-4S cluster through the formation of PTN. Other proteins like TOM 71, Putative flavin-dependent monooxygenase, isocitrate dehydrogenase etc. proteins can also be nitrated. The activities of these enzymes are lost due to PTN [16]. MnSOD is another enzyme whose activity can be inhibited by PTN [92]. MnSOD inhibits the formation of peroxynitrite by dismutating superoxide. This inactivation of MNSOD leads to nitration of other key proteins of mitochondria [93]. This nitration compromises antioxidant defenses which may lead to apoptosis and structural alteration of protein [94]. Some studies indicate the presence of a denitration mechanism within the mitochondria which is oxygen tension-dependent but protease-independent [95]. Total PTN increased in mitochondria when whole organs were exposed to ischemic hypoxia. The denitration of several mitochondrial enzymes during the hypoxic-anoxic phase may help to sustain mitochondrial ATP production and an increased antioxidant capacity, which is important for the prevention of metabolic failure and cell death during oxygen deprivation [90]. Such reports lead to the hypothesis that within mitochondria protein modification by nitration is reversible which is maintained by concerted effect of a nitration-denitration system [96].
Elucidating the denitration process in vivo is still a challenge for the scientists. For decades PTN was thought to be a stable post-translational modification but in several studies with different proteins like HDL-containing plasma lipoprotein fractions [97], isolated platelets [98], activated macrophages [99] and calmodulin, the presence of a selective denitration mechanism was observed in vitro. Denitration is a selective process of like nitrated COX-1 protein isolated from macrophages, human and murine endothelial cells, and different tissue samples is one of the substrate for denitration. Denitration of nitrated COX-1 was quantified and validated by HPLC (High-Performance Liquid Chromatography) with electrochemical detection. This denitration activity was time and concentration dependent and it was inactivated by heating and proteolysis suggesting the association of a protein [100]. Another study showed that inspite of a lack of a nitratase activity albumin can remove ONOO- from the blood circulation using a ca2+-dependent pathway with the concomitant increase of nitrated albumin [97,101]. A Ca2+-dependent denitration process was also seen in HDL and LDL -containing samples of heart and brain homogenates where nitrate ion concentration increased stoichiometrically with the denitration process [97]. Another calcium-dependent denitration system was seen in freshly isolated platelets in which nitro group was directly removed from tyrosine residues without being reduced to aminotyrosine (NH2-TYR) [102]. Reduction of nitrotyrosine to aminotyrosine is purely a chemical process mediated by agents like DTT (Dithiothreitol) and Fe3+-containing heme. Proteins like hemoglobin and myoglobin are also capable of catalyzing such reduction reaction [103]. A very recent study with Saccharomyces cerevisiae showed that 14 nitrated proteins involved in signal transduction pathway as well as the cellular functions also have an important role in mating signaling pathway. The functional interaction of these 14 nitroproteins is revealed by network analysis and other bioinformatic tools. One of the important protein for mating signaling, YPD1, is activated by nitration which helps it to stimulate pheromones. The regulation of such a protein between its active and inactive state is assumed to be mediated by a nitration-denitration system [15]. Some studies indicate the presence of putative denitrase in vivo which can maintain the dynamics of protein tyrosine nitration and denitration [104-106]. But still the actual mechanism of such denitration is not clear.
One of the low-yield biological processes is PTN. Thus highly selective and specific methods are needed to quantify 3-nitrotyrosine [v]. In last year’s many methods are developed to detect either circulating free form of 3-nitrotyrosine or the form which is released after protein hydrolysis [108]. Depending on the disease and in the level of free nitrotyrosine and tyrosine-nitrated proteins are dependent on the type of disease as well as the tissue of interest [109]. The methods are not error free. Analysis of 3-nitrotyrosine is the main drawback of these methods, for example concentration of individual nitrated protein [110].
It is one of the effective methods to identify the PTN. The extensively used method to study nitroproteome is 2DGE followed by western blotting [95]. The most common antibody for immunobloting is the clone 1A6monoclonal antibody [111]. Other antibodies with high specificity for 3-nitrotyrosine are also used to detect nitrated proteins [112]. But non-specific binding is one of the major problems. To solve this problem reducing agents-mediated reduction of nitrotyrosine is applied [95]. But there is another problem that sodium dithionite is sensitive to atmospheric oxygen so less amount of proteins are only reduced [92]. For that sensitivity of sodium dithionite, aminotyrosine again back to nitrotyrosine due to the reoxidation. This event may lead to weak signal of 3-nitrotyrosine on dithionite-treated membranes [111]. Low abundance of 3-nitrotyrosine in protein, extreme pI, size, solubility of proteins are the important facors which are related to the success of 2DGE approaches.
Several immunehistochemistry are widely used to detect NO2-TYR. Different monoclonal and polyclonal antibodies are used in these studies to precipitate antigen-antibody complex. These antibodies are commercially available. But the antigen-antibody complex is not formed due to some reasons like pretreatment of the samples with reducing agents (e.g, dithionite), excess amount of NO2-TYR, treatment with nitrated bovine serum albumin [108].
Though low abundance of pure samples and week sensitivity are the problems but still Ultraviolet–Visible Spectroscopy is used to quantify the free NO2-TYR as well as the NO2-TYR which are present in peptides and proteins. Due to the change in pKa value of nitrotyrosine, the absorbance spectra can also be changed. Thus, the concentration of nitrotyrosine is quantified at the absorbance of 430 nm [113].
In several studies nitro-tyrosine is quantified by HPLC. Enzymatic or acid hydrolysis is required to prepare the samples [114]. But the major problem of this technique is verification. Through NO2-TYR is stable to acid hydrolysis [115] but it is reported that NO2-TYR is converted to NH2-TYR during acid hydrolysis.
Tyrosine and nitro-tyrosine, both are detected by using UV detector (absorbance maxima at 280 nm at pH 3.5) which provides an output signal proportional to the absorption. More selective identification of NO2-TYR is achieved at the absorption spectra of 365 nm at pH 3.5. In this technique acid hydrolysis is also done [108]. A more sensitive electrochemical detector is also used to quantify the concentration of protein tyrosine nitration and denitration [100].
NO2-TYR can be detected by using fluorescence detector but it is not a fluorescent compound. At first fluorescent product is prepared by the reaction of nitrotyrosine and derivatizing reagent such as o-phthalaldehyde (OPA) or 7-fluoro-4-nitrobenzo-2-oxa-1,3-diazole (NBD-F) to quantify the NO2-TYR [108].
Nitrotyrosine can be detected by using gas chromatography. In this technique tetranitromethane treated sample is used and incubated in vitro. In this technique a thermal energy analyzer is used with gas chromatography. Minimum 0.5 ng NO2-TYR is detected per injection. GC–MS (Gas Chromatography-Mass Spectroscopy) is used to identify and isolate nitrated proteins of urinary metabolites as 3-nitro-4-hydroxyphenyllactic acid and 3-nitro-4-hydroxyphenylacetic acid [108]. But for GC-MS technique the proteins are not directly injected because amino acids are not volatile and thermally stable. To solve this problem functional group of PTN is modified to increase the thermal stability and volatility [107].
Nitrotyrosine can be detected by using mass spectroscopy equipped with standard nitrogen laser (337 nm) and delayed extraction optics [116]. In some studies nitrotyrosine is detected by very sensitive GC-MS method. In this technique protein are hydrolyzed by acid and then aminotyrosine is detected by GC-MS. Heptafluorobutyric anhydride (HFBA) is used in this technique. HFBA reacts with different functional groups like amines and hydroxyls. At first the carboxylic group of 3-nitrotyrosine is converted to n-propyl ester and then the derivatives reacts with HFBA and form a compound which is detected by negative ion chemical ionization mass spectrometry (NICI–MS). NO2-TYR of heart proteins of rat was quantified by this technique. NO2-TYR and TYR was diluted with the isotope of 13C-labeled analogues of these two molecules to quantify the level of tyrosine and nitrotyrosine [117]. Nowadays a developed mass spectroscopy approach LS-MS/MS is used to get the more accurate profile of PTN. Nanoflow LC-MS/MS (Liquid chromatography-tandem mass spectroscopy) was used to identify the peptides and proteins from different denitrase-containing fractions. The fractions were required to enrich on a DEAE-Sepharose anion exchange column because the amount of nitroprotein is very less in vivo [100].
Bioinformatic tools have a great impact on genomics and proteomics studies. Different tools are used in proteomics to identify the proteins, their structure and functions. One of the softwares TurboSEQUEST, is used for identification of nitrated protein [118]. Scaffold software which is used in these tools helps to compute protein and peptide probability for nitration [119]. The proteins having more than 90% probability are accepted [15]. During identification of the homologs of different proteins by MS-BLAST the de novo sequences are correctly tagged. But the sequence tagging is error-prone. To solve this problem another prevalently used software SPIDER is used where both the errors, de novo sequencing and homology mutations are taken into account [120].
Tyrosine residues which are present in loop are more susceptible for nitration. Thus secondary structure of protein is important factor to predict [121]. Secondary structure of proteins can be determined by using different algorithms e.g. PSIPRED (Software for Protein Identification from Sequence Tags with De Novo Sequencing Error), JNET, PROF etc. [122-126]. One of the best methods to predict the protein structure is neutral network. The ability of PSI-BLAST is exploited by PSIPRED [122]. A more strong alignment profile is built by using PSI-BLAST. Compared to the conventional pairwise sequence searching methods, the more distant similarities are also included in this alignment profile [124]. JPred is used to predict the secondary structure of proteins. JPred4 is the latest version of JPred where JNet algorithm is used. JNet algorithm is one of the most error-free methods to predict the secondary structure of protein. One of the major characteristic of JPred4 is higher accuracy. All the three states (-strand, -coil and -helix) of secondary structure of proteins can be predicted by JPred4 with the 82% of accuracy. This accuracy is raised to 90% while solvent accessibility is predicted [127]. A useful software for characterizing the PTN site is GPS-YNO2 having 76.51%, accuracy, 50.09% sensitivity and 80.18% specificity from the leave-one-out validation [128].
The tyrosine residues which are exposed to solvent phase only available for nitration [121]. Thus Solvent accessibility is another important parameter to predict the protein structure because this parameter is related to the packing of amino acid residues and spatial arrangement during the protein folding. This parameter is also closely related to functions of proteins, protein–ligand interactions, protein–protein interactions [129]. Solvent accessibility is efficiently predicted by SANN, a nearest neighbor method.
Another bioinformatics method used to determine the role and interaction of a protein in certain protein pool is protein networking [130]. There are two major network models for proteomics- PIN (protein interaction network) and PSN (protein-signaling network). PIN is used in proteomics where protein-protein bindings are involved whereas PSN is used to study the mainly post-translational modification like tyrosine nitration. There are several databases for protein interactions e.g. DIP, BIND, MIPS, MINT etc. DIP (The Database Interacting Proteins) contains only experimentally derived data of protein-protein interaction. It is one of the largest collection of publicly accessible protein-protein interaction information. The DIP data model consists of binary protein interactions along with information on each protein, the method used to determine the interaction, and a set of publication references to support the record. JDIP is stronger software which has the higher visualization power and decreases the problems with the increasing complexity and size of the data available in DIP [131]. DIP allows queries by proteins, BLAST, protein sequence motifs, and by journal article. An analysis of the confidence level of interaction in the DIP database has been published, and two types of confidence of interaction scores are available: PVM (paralogous verification method), assigns a higher reliability score to an interaction whose paralogs are also seen to interact in DIP. Another score is EPR (Expression Profile Reliability) has the ability to deem a set of protein-protein interactions more reliable if it has a similar expression profile as a higher-quality subset of DIP. But the score is only calculated for budding yeast because large amount of external information currently only available for budding yeast [132]. BIND (Biomolecular Interaction Network Database) is the one of the largest collection of freely available information about pairwise molecular interactions and complexes. Information about the interactions between two biological ‘objects’ like DNA, RNA, protein, gene, molecular complexes, small molecules are stored by BIND and the probable pathways are also stored by BIND. The specification of BIND data is available as XML DTD and ASN.1 [133]. MINT (The Molecular INTeraction database) contains data on the functional interaction between proteins. Mainly this consists of direct protein-protein interactions, but also includes indirect and genetic interaction. MINT can store enzymatic modifications and information on binding domains and kinetics [134]. MIPS mammalian protein–protein interaction database (MPPI) is a large collection of the data of high-quality experimental protein-protein interaction in mammals. Only experimental data are included here to maintain the quality. MySQL is the database where all these data are stored and these data are accessible through Perl CGI scripts, a web interface. Protein with all its neighbors is graphically represented by MIPS. It is a special feature of MIPS. The entire dataset can be downloaded in the standard format of PSI-MI for detail analysis. Different additional informations such as the experimental technique which are used, PubMed reference, probable sites for binding, and also the functional role of the protein-protein interaction are provided by MIPS [135].
Increase in the data content is a huge problem for visualization. One of the solutions of the problem is Cytoscape. Cytoscape is a stronger freely available, open-source java-based network visualization and analysis tool. Due to this ability non-programmers are allowed to plot the graphical images onto nodes and introduction of spreadsheet-like equations make it a stronger tool for integrating and visualizing complex datasets [136] (Figure 3a and b).
These software’s can be used to detect protein tyrosine nitration and denitration system. It will be easier to characterize proteins by using different bioinformatics tools. If several enzymes are involved in denitration system then the best choice to establish the probable pathway is protein networking. The secondary structure and Solvent accessibility are also very important to understand the structure and stability of the protein. The use of bioinformatics tools can be very helpful to characterize and establish the probable pathway of denitration system.
Low abundance of nitroprotein is the major problem to analyze 3-Nitrotyrosine as well as the dentration system. Some data suggest that denitration is an enzymatic process. But till now denitrase enzyme is not purified. So the purification is the challenge for the scientists. Our hypothesis is that to perform the western blot of cell free extract (CFE) and then treat the membrane of western blot with fractionated cell free extract (CFE). 2DGE and LC-MS/MS will be performed of the fraction which shows the highest denitration activity (Figure 4). If the dentration system is associated with several proteins, then protein networking has to be done (Figure 5).
The study of PTN has gone through more than 50 years. Though it is a stable post translational modification but recent studies indicate that the process is reversible. But the in vivo role of PTN is still not very clear. There is almost no strong evidence of reversion of nitrated proteins. The biological significance of PTN is the study of interest in recent times. PTN is not only a simple in vivo one step reaction but it is related to signal transduction. But in human the altering of protein function by PTN is a fundamental fact that has been proved its drastic effects in neurodegenerative diseases. The main cause of neurodegenerative diseases is the tangle formation of neuroproteins. This event makes the proteins inactive. Denitration on the other hand is a mechanism that has been quite intriguing, foraying our perspective into a process that can alter nitration and thus may pave our way to a whole new world of perspectives where neurodegenerative diseases can be cured. Till now very few medicines are available for the remedy of these diseases. But these medicines have side effects. But the probability of using “denitrase” enzyme as a therapeutic medicine for the neurodegenerative diseases will be an interesting aspect to study. So the nitration–denitration pathway is very important to fully understand. If the nitration-denitration system is discovered in the microbes, a less complex cellular system then it will be the model for the higher eukaryotes and remedy for the neurodegenerative diseases because it is hypothesized that ‘Denitrase’ will prevent the oligomer formation [11,15,17,104-106] (Figure 6). But still isolation and application of ‘Denitrase’ is a challenge for scientist.
The authors acknowledge the University of North Bengal for providing essential infrastructure to carry out this research.
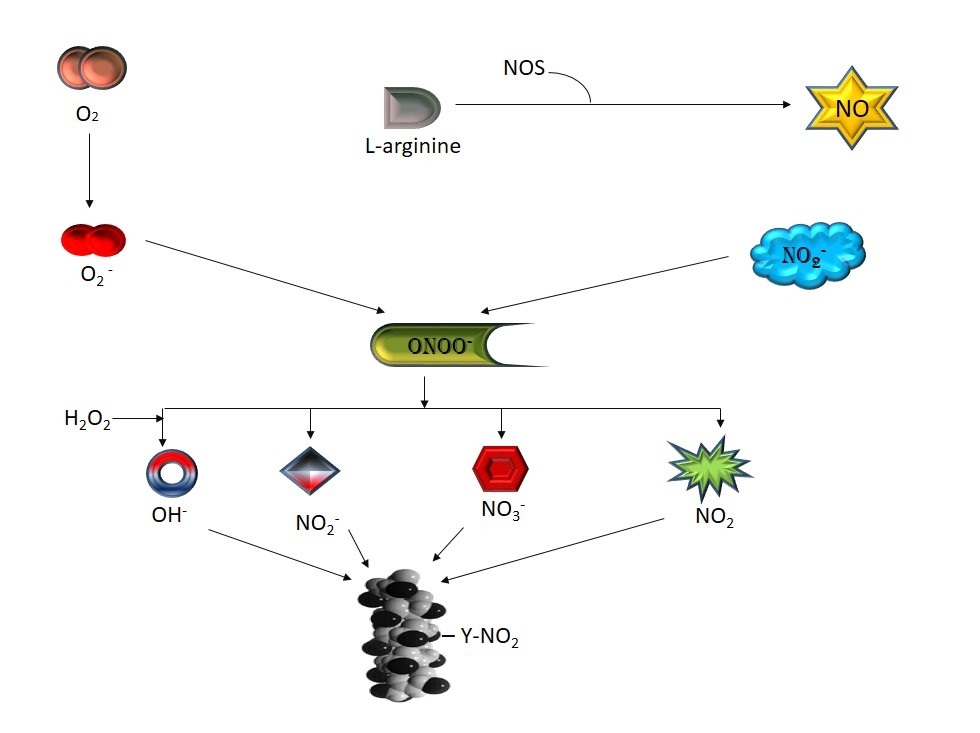 |
Figure 1: Formation of different reactive species from peroxynitrite |
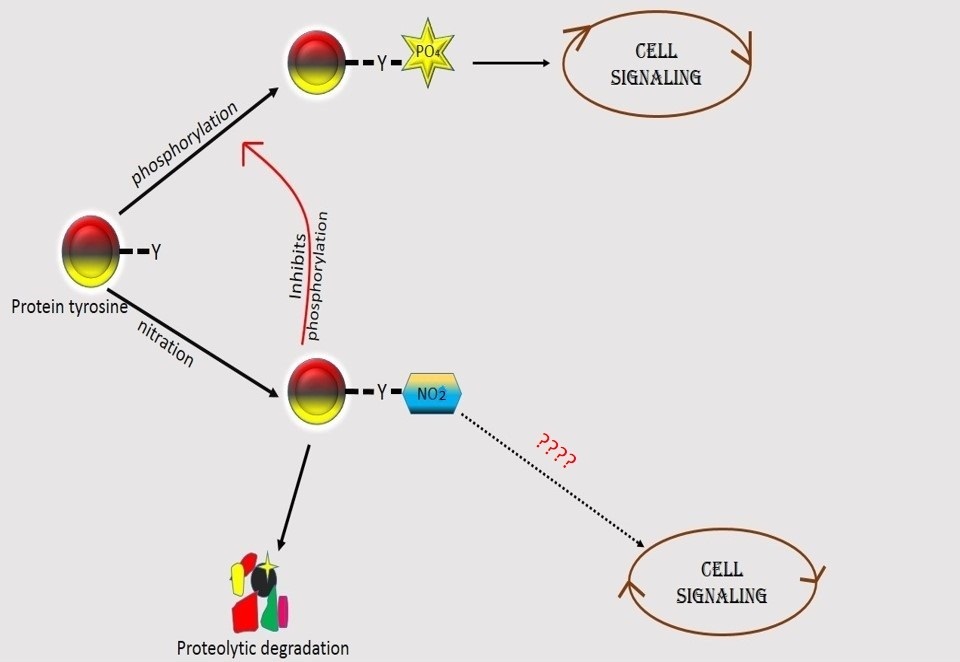 |
Figure 2: Schematic diagram of the role of nutrition and phosphorylation on tyrosine |
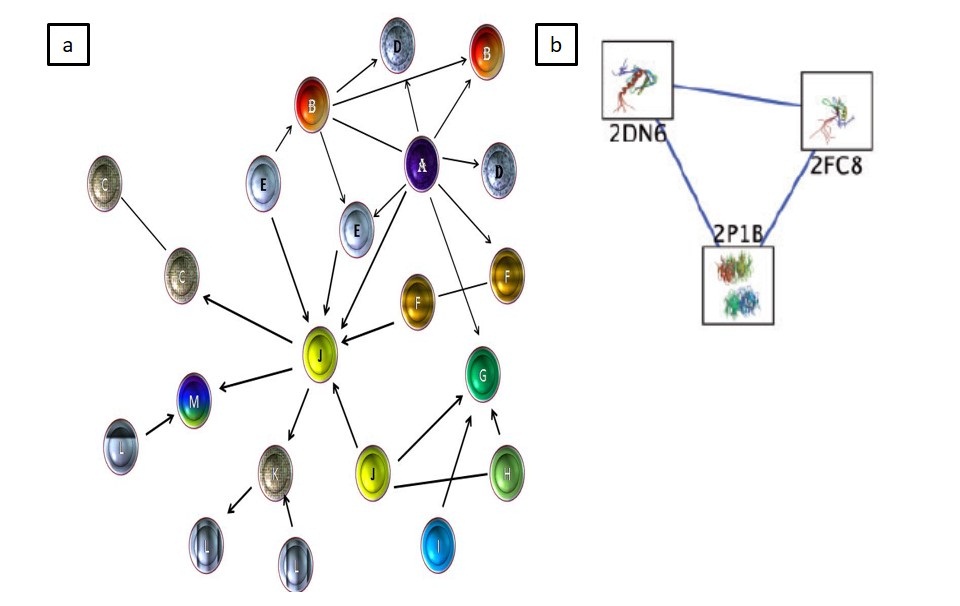 |
Figure 3: (a) A model of protein networking visualization by cytoscape (b) protein–protein interaction network visualization by cytoscape (adapted from Smooth ME et al. 2011) |
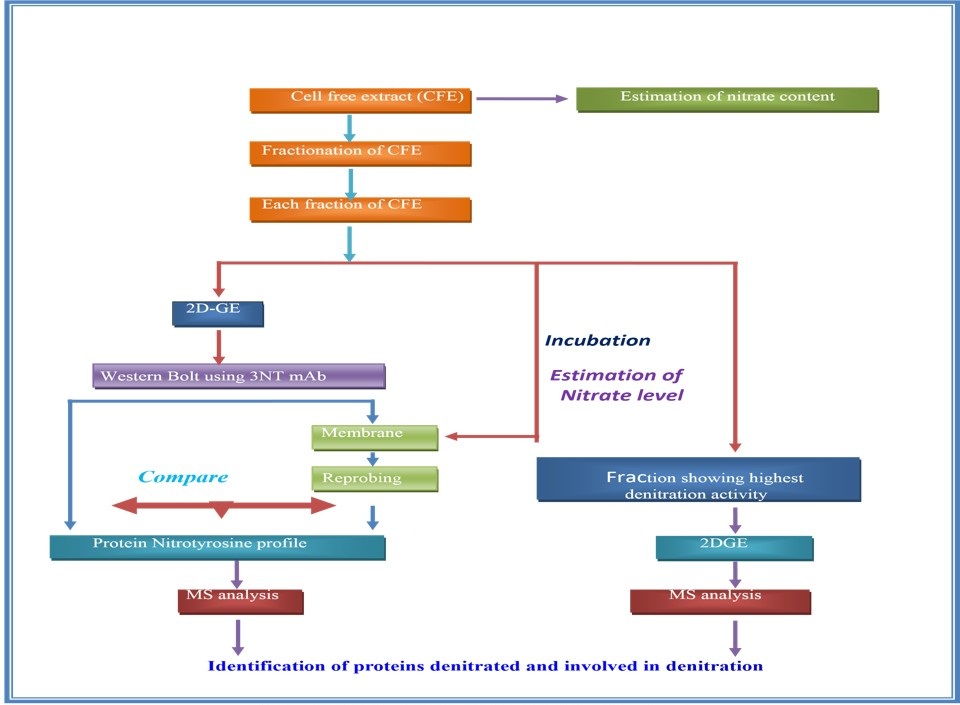 |
Figure 4: Probable method to detect “DENITRACE” enzyme |
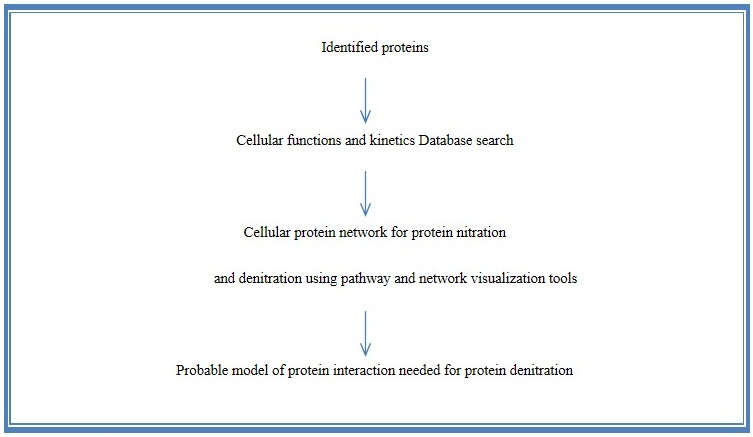 |
Figure 5: Approach for determining the cellular protein network for denitration |
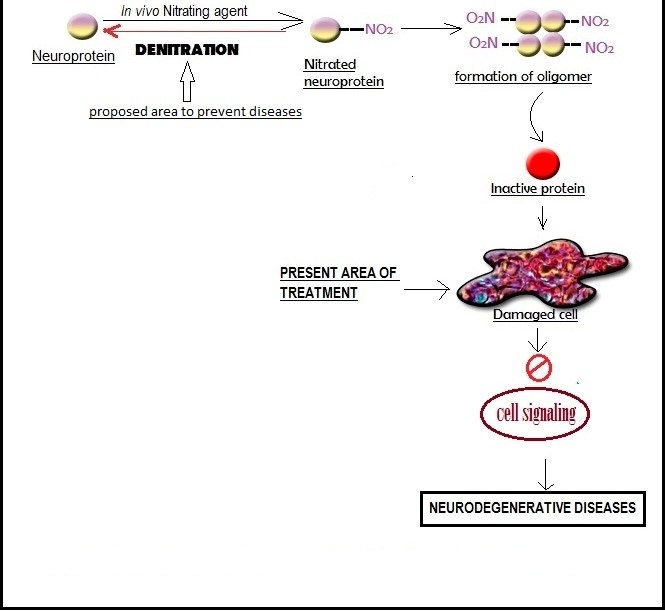 |
Figure 6: Future perspective to prevent the Neurodegenerative disease |






































