Top Links
Journal of Computational Systems Biology
ISSN: 2455-7625
Synteny Based Analysis to Proffer a Non-drug Resistant Target Against Porphyromonas gingivalis
Copyright: © 2014 Ashok Kumar. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Related article at Pubmed, Google Scholar
Porphyromonas gingivalis is an anerobic, gram negative, bacterium which is a natural inhabitant of oral microbiome. Under unhealthy oral hygiene conditions the bacterium becomes highly destructive (photobiont) and leads to periodontitis (inflammation of periodontium). Several drugs like chlorhexidine, monocycline effectively reduce the viability of biofilms formed by Porphyromonas gingivalis but, it is impossible to destroy biofilms completely. As the bacterium evolves continuously it develops drug resistance due to events like horizontal gene transfer and its biofilm mode of living. Therefore no drug is very efficient in treating Periodontitis. We have done an Insilco Synteny based comparative analysis to find a druggable target which remains unchanged in various Porphyromonas gingivalis strains. Dipeptidylaminopeptidase has been identified one such potential target which is responsible for virulence in Porphyromonas gingivalis, and is functionally and structurally conserved. Therefore, according to best knowledge of authors a drug developed against this target will be effective in treating periodontitis caused by all evolved forms of bacterium.
Keywords: Porphyromonas gingivalis; Periodontitis; Drug resistance; Synteny
Abbreviations: DPP IV: Dipeptidylaminopeptidase IV; BLAST: Basic Local Alignment Search Tool; NCBI : National Center for Biotechnology Information; RMSD: Root Mean Square Deviation; ATCC: American Type Culture Collection; MSA: Multiple Sequence alignment
Periodontitis is inflammatory diseases caused by peridontopathic bacteria, majorly acquires gingival crevice, often causing gums to shrink and loosening of teeth. It is apparently caused by specific gram negative anaerobic bacteria [1], Porphyromonas gingivalis, Treponema denticola, and Tannerella (formerly Bacteroides) forsythia more commonly termed as the ''red complex''[2,3]. Porphyromonas gingivalis is a natural member of more than 500 bacterial species that live in oral microbiome, yet it becomes immensely catastrophic (termed pathobiont) and proliferates to high cell numbers in periodontal lesions: this is credited to its arsenal of specialized virulence factors. Gingival epithelial cells, the outer lining of the gingival mucosa, which function as an innate part of the innate immune system, are among the first host cells colonized by Porphyromonas gingivalis [4]. Treponema denticola leads to necrotizing ulcerative gingivitis after getting adhered to fibroblasts, epithelial cells, as well as to extracellular matrix components present in periodontal tissues. It posses outer-sheath-associated peptidases, chymotrypsin-like and trypsin-like proteinases, hemolytic and hemagglutinating activities, adhesins as virulence factors which aid in early periodontitis [5]. T.forsythia is another anaerobic, Gram-negative bacterium isolated from the gingival sulci and periodontal pockets of patients with periodontitis [3]. Several T.forsythia virulence factors have been identified [6], including, trypsin-like [7] and PrtH proteases [8], sialidases SiaH [9] and NanH [10], a leucine-rich repeat cell-surface-associated and secreted protein BspA [11], an apoptosis-inducing activity [12], alpha-D-glucosidase and N-acetyl-beta-glucosaminidase [13], a hemagglutinin [14], components of the bacterial S-layer [15].
Within group of red complex, P. gingivalis and T. denticola are frequently found together in diseased sites [3], but T. forsythia, may not co localize spatially with them [16,17]. Also T. denticola do not form homotypic biofilms. It is P. gingivalis which acted as an initial colonizer of the substratum, enabling subsequent incorporation of T. denticola [18,19]. Gingipains (P. gingivalis) and dentilisin (T. denticola) have happened to be involved in the co aggregation [19,20]. Due to the fastidious growth requirement for culturing T. forsythia and the genetic manipulations of this organism are difficult to perform, this organisms is understudied [21].Among the species of red complex, P. gingivalis has happened to be a major pathogen, with well-defined virulence factors including cell surface located proteolytic enzymes and adhesins (gingipains) [22,23]. Recent studies have focused on the pathogenic traits of the Porphyromonas gingivalis. A pioneering work by scientist suggests that Porphyromonas gingivalis produces an array of potential virulence factors viz capsule, outer membrane protein and gingipains etc. It possesses strong proteolytic activity, which is considered to be associated with tissue destruction. It cleaves epithelial junction proteins, which suggests that it invades deep into tissue via paracellular pathway and thereby destroying the connective tissue. Identified proteases are Arg-specific and Lys-specific cysteine proteases, DPPIV [24], prolyltripeptidyl peptidase (PtpA), and others [25].
There are several ways of treating periodontitis, non-surgical methods like (scaling and root planning), surgical methods (flap surgery, bone and tissue grafts) medication is used with non-surgical treatment but, it has been seen that Porphyromonas gingivalis biofilms persist even after treatment with chlorhexidine (chlorinated phenolic disinfectant used commonly in mouthwash for its action against bacteria) [26] and other known antibiotics also. The bacterium continuously evolves since the biofilm it forms with other bacteria provides an ideal niche for the exchange of extrachromosomal DNA which encodes for resistance to antimicrobial agents and therefore making it difficult to treat bacteria [27]. It is known that certain residues that perform key functional and structural roles tend to be preserved by natural selection. In this regard discovering genome level similarity based on modern approach like Synteny Based Comparative Genomics is serving us to find precisely the essential genes which are conserved and play a role in intensification of bacterial infection as well as in promoting drug resistance in case of periodontitis and therefore, can be potential drug targets.
NCBI Genome was used for genome selection of Porphyromonas gingivalis. Only the genomes that were completely sequenced (Porphyromonas gingivalis ATCC 33277 and Porphyromonas gingivalis TDC60) were selected for carrying comparative genome analysis. According to literature and database availability of complete genome information, we opted for Porphyromonas gingivalis W83 as a reference genome.
Microscope server (www.genoscope.cns.fr/agc/microscope/home) [28] was used to carry out the synteny analysis between the three fully sequenced genomes of Porphyromonas gingivalis. Firstly the reference genome was selected and loaded into the browser and then the other two strains (Porphyromonas gingivalis ATCC 33277 and Porphyromonas gingivalis TDC60) were selected. Homologous synteny blocks were identified based on Microscope search. The list of the conserved synteny blocks was downloaded in the form of MS-Excel data.
From the synteny data obtained, variable and non- variable genes was filtered based on the observation that a particular gene from the comparable genomes corresponded to only a single gene in the reference genome also, such genes were taken under non- variable genes and the conserved blocks that corresponded to more than one gene in the compared genomes were kept under variable genes. Then, a literature study was performed using PubMed to determine the major virulent factors present in Porphyromonas gingivalis. These genes was then searched in non-variable genes and a list of virulent factors were made and each of which was individually studied to find a factor or a potential target that was conserved in all the strains and had role in increasing the pathogenic potential of Porphyromonas gingivalis.
Insilico characterization of target was done by retrieving the nucleotide and amino acid sequences from NCBI. MSA using MUSCLE [29] was done for protein as well as nucleotide sequences.
The selected target was validated performing a database search using BLAST [30] to see whether the target was specific to the bacterium or it was homologous to some human protein. After BLAST results, conserved domains were predicted using InterPro [31] to see structurally and functionally similar domains in the target sequences from three different strains of Porphyromonas gingivalis.
Only the completely sequenced genomes of Porphyromonas gingivalis were selected. Porphyromonas gingivalis W83 was taken as a reference genome and was compared with the other two completely sequenced genomes of Porphyromonas gingivalis ATCC 33277, and Porphyromonas gingivalis TDC60. Results from Microscope server showed conserved synteny blocks between the reference and other two strains. Colored boxes in the results represent synteny conservations in at least one organism of the related taxon (Figure 1).
Literature studies suggested that peptidases in Porphyromonas gingivalis were the major reason for its virulence and therefore from the list of non-variable genes (Table 1), another list of all the peptidases was made and analyzed further and out of which DPP IV was selected as a potential target (Table 2).
Nucleotide sequences of the three DPP IV genes from three different strains were analyzed by performing MSA by MUSCLE. The percent identity matrix has identity of 61% between the sequences and hence was found to be homologous to each other. Percent identity matrix of amino acid sequences revealed identity of more than 30% hence was homologous. Amino acid sequences retrieved from NCBI had shown that the amino acid number varied from first sequenced to recently sequenced strains Porphyromonas gingivalis W83 had 732 amino acids [32], Porphyromonas gingivalis ATCC 33277 had 723 amino acids [33] and Porphyromonas gingivalis TDC60 had 709 amino acids [34], respectively. This variation in amino acid numbers could be due to evolution of bacterium and would have led to some loss or gain in function.
In order to make sure that Porphyromonas gingivalis DPP IV is specific and is not found in humans, BLAST database search was conducted against non- redundant protein database and results gave no significant similarity with any human protein for three DPP IV sequences. Therefore we conclude that the target selected was specific to Porphyromonas gingivalis.
InterPro gave the two common conserved domains for three DPP IV sequences. IPR029058 (a region in DPP IV predicted as Alpha/Beta hydrolase fold; this structure is common to a number of hydrolytic enzymes of widely differing phyologenetic origin) and IPR002469 [a region in DPP IV as belonging to serine peptidase family (S9)]. This led us to believe that even though the sequence is changing, functionally and structurally important regions are still conserved. Therefore, a drug made against this target will remain effective for all Porphyromonas gingivalis strains and since DPP IV is highly conserved it will not show resistance to the drug.
Secondary structures was predicted using PHD [35] showed that number of alpha helices was increasing in newly sequenced bacteria hence making DPPIV a more stable structure (Table 3). Tertiary structures were generated using SWISS MODEL and were then visualized in Chimera, all the three DPP IV structures predicted were found to be homodimers.
Verify3D [36] validated structures generated by SWISS MODEL, all structures obtained profile scored greater than 0.2. The structure was also validated by generating Ramachandran plots of the three models and by calculating RMSD values [37] for the superimposed structures in chimera (Table 4). Maximum residues in Ramachandran plot lied in allowed regions and RMSDs obtained using both Smith Waterman and Needleman Wunch algorithm were less than 2 angstrom.
The current study illustrates an emerging field of comparative genomics i.e. synteny based analysis which allows us unmask the conserved gene content and gene order. Identifying Synteny blocks and aligning whole genome both rely on markers across the genomes long enough to make their conservation statistically significant [38]. Till date many conventional therapies have been used in treating periodontitis but eventually fail due its complex multi-factorial etiology [39]. Therapies include mechanical removal of bacterial deposits and calculus from subgingival environment or performing rooting and scaling followed by use of antibacterial agents mainly chlorhexidine, tetracycline, doxycycline and minocycline but local delivery is cost effective. They bind to tooth and reduce bacterial count and dental plaque, out of which chlorhexidine is a gold standard in dentistry [40,41]. Since P.gingivalis colonizes first to the tooth surface followed by synergy association with T. denticola in establishing biofilms and with T.forsythia, it is assumed that if a target is chosen from P.gingivalis it will strongly effect the association which the organisms have with other two anaerobes. According to the literature, many protease targets have been reported which accounts for strong proteolytic activity due to which P. gingivalis invade into deeper tissues [4]. With the aid of synteny based study, we are able to deduce one such potential druggable target i.e. DPP1V protease which is involved majorly in pathogenesis of bacterium. This target is species specific and structurally and functionally conserved in all the known three strains of P. gingivalis. To wrap up if drug is made against this target of P. gingivalis which happened to be primary colonizer, it would staunch enough treat to periodontitis effectively.
Synteny based analysis of P. gingivalis has led to testimony of diverse proteases that can be used for potential target for drug design. As various proteases have been noted to play a substantial role in pathogenesis of periodontitis, a systematic approach to develop drug will be propitious for treatment. In this study DPP1V has found to play a decisive role in breaching deeper tissues and biofilm formation which happens to be an imperative factor in periodontitis. It is expected that if drug is developed against this will be roseate in cure.
Authors are thankful to Vice Chancellor of Panjab University, Chandigarh, and Coordinator of Centre for Systems biology and Bioinformatics for providing all the facilities to carry out the present work. We acknowledge the free softwares, tools and web servers used in this present study (NCBI genome, Microscope server, MUSCLE, BLAST, InterPro, SWISS model, PHD, Chimera).
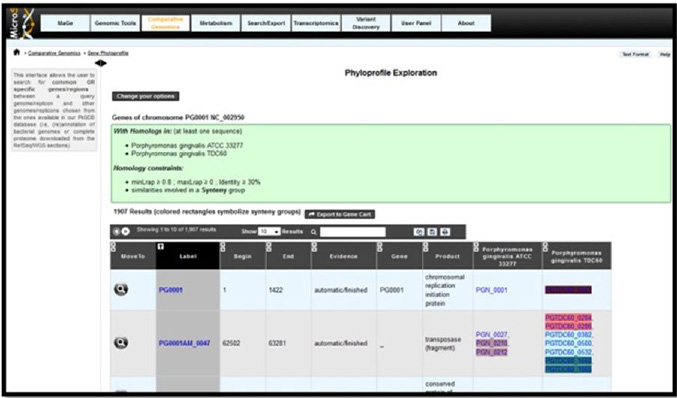 |
| Figure 1: Microscope Synteny homepage |
| Begin | End | Porphyromonas gingivalis W83 | Product | Porphyromonas gingivalis ATCC 33277 | Porphyromonas gingivalis TDC60 |
| 1 | 1422 | PG0001 | Chromosomal replication initiation protein | PGN_0001 | PGTDC60_0001 |
| 1435 | 2007 | PG0002 | Hexapeptidetransferase | PGN_0002 | PGTDC60_0002 |
| 2009 | 3028 | PG0003 | Hypothetical protein | PGN_0003 | PGTDC60_0003 |
| 3108 | 3812 | PG0004 | NAD-dependent deacetylase | PGN_0004 | PGTDC60_0004 |
| 3887 | 5041 | PG0005 | Hypothetical protein | PGN_0005 | PGTDC60_0005 |
| 5094 | 6422 | PG0006 | Mate efflux family protein | PGN_0006 | PGTDC60_0006 |
| 11634 | 14645 | PG0011 | gycosyl hydrolase | PGN_0009 | PGTDC60_0011 |
| 11693 | 15721 | PG0012 | L-threonine O-3 phosphate decarboxylase | PGN_0010 | PGTDC60_0012 |
| 15731 | 16501 | PG0013 | Hypothetical protein | PGN_0011 | PGTDC60_0013 |
A list of non variables genes was obtained. |
|||||
| S.no | Peptidases in Porphyromonas gingivalis | Corresponding genes ids |
| 1 | Signal peptidase like protein | PG2000, PGN_1945, PGTDC60_0273 |
| 2 | Signal Peptidase 1 | PG2001 , PGN_1946, PGTDC60_0275 |
| 3 | M16 family peptidase | PG0088, PGN_1946, PGTDC60_0367 |
| 4 | Aminoacyl-histidinedipeptidase | PG0137, PGN_0250, PGTDC60_0414 |
| 5 | EndopeptidasePepO | PG0159, PGN_0271, PGTDC60_0510 |
| 6 | Zinc carboxypeptidase | PG0232, PGN_0335, PGTDC60_0510 |
| 7 | M49 family peptidase | PG0317, PGN_1645, PGTDC60_1437 |
| 8 | Dipeptidylaminopeptidase IV | PG0503, PGN_1469, PGTDC_1620 |
| 9 | Peptidase T | PG0445, PGN_1517, PGTDC60_1565 |
| 10 | Prolyloligopeptidase | PG0724, PGN_1776, PGTDC60_18845 |
| 11 | Peptidyl-dipeptidaseDcp | PG1789, PGN_0756, PGTDC60_0105 |
| 12 | M24 family peptidase | PG0889, PGN_1050, PGTDC60_0186 |
| 13 | Cysteine peptidase | PG1788, PGN_1777, PGTDC60_0106 |
From the list of non- variable genes after literature study peptidases were separated into a different list |
||
| Strains | Alpha helices | Extended strands | Random Coils |
| Porphyromonas gingivalis W83 | 14% | 33.33% | 52.19% |
| Porphyromonas gingivalis ATCC 33277 | 25.73% | 28.77% | 45.50% |
| Porphyromonas gingivalis TDC 60 | 26.52% | 29.66% | 44.43% |
Increase in the number of alpha helices and decrease in the number of, extended strands and random coils are seen. |
|||
| Algorithm used | RMSD Value W83 with ATCC 33277 | RMSD Value W83 with ATCC 33277 | |
| Needleman Wunch Algorithm | 1.221 | 1.238 | |
| Smith- Waterman Algorithm | 1.221 | 1.238 | |
RMSD values obtained were less than 0.2 angstroms |
|||






































