Top Links
Journal of Cell Biology and Histology
ISSN: 2638-082X
Potential Molecular Players Against to SARS-CoV-2: A Glyco-Perspective
Copyright: © 2021 Tugrul B. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Related article at Pubmed, Google Scholar
COVID-19 is caused by SARS-CoV-2 and is associated with the increasing number of cases and deaths and has been declared a pandemic by WHO. The treatment strategy focused on preventing the S protein from binding and penetrating the ACE-2 receptor. In addition, S protein-related molecules such as peptides, blockers, and inhibitors that inhibit S protein cleavage are considered candidate therapeutics. However, the major hurdle in the development of anti-viral therapy appears to be glycosylated components of a virus. Host-derived glycosylated molecules of a particular virus can protect them from immune attack and increase their virulence with masking effects. In this review, various aspects of glycans regarding virus-host interactions and new therapeutic concepts on the glycosylation machinery are discussed.
Keywords:SARS-CoV-2; S protein; Glycosylation; Furin; TMPRSS2
List of abbreviations:ACE-2: Angiotensin Converting Enzyme 2 Receptor; ARDS: Acute Respiratory Distress Syndrome; CD: Connector Domain; CH: Central Helix; COVID-19: Coronavirus Disease-19; CT: Cytoplasmic Tail; CTD: C-terminal Domain; E: Envelope; ER: Endoplasmic Reticulum; ERGIC: Endoplasmic Reticulum-Golgi Intermediate Compartment; FP: Fusion Peptide; Gals: Galectins; MBL: Mannose-Binding Lectin; HIV-1: Human Immunodeficiency Virus Type 1; HR1/2: Heptad Repeat 1/2; HR1: Heptad Repeat 1; HR2: Heptad Peptid 2; IFP: Internal Fusion Peptide; M: Membrane; MERS-CoV: Middle East Respiratory Syndrome Coronavirus; MHC: Major Histocompatibility Complex; N: Nucleocapsid; NSPs: Nonstructural Proteins; NTD: N-terminal Domain; ORFs: Open Reading Frames; PTMs: Post Translational Modifications; RBD: Receptor-Binding Domain; RdRp: RNA Polymerase; S: Spike Protein; S1: Subunit 1 of Spike Protein; S2: Subunit 2 of Spike Protein; SARS-CoV: Severe Acute Respiratory Syndrome Coronavirus; SARS-CoV 2: Severe Acute Respiratory Syndrome Coronavirus 2; SP: Signal Peptide; TM: Transmembrane; TMPRSS2: Transmembrane Protease Serine 2; WHO: World Health Organization
Most patients who are suffering from COVID 19 caused by the SARS-CoV2 virus, which is included in the betacoronavirus genus, show mild or moderate symptoms. 20% of patients have a phenotype caused by other coronaviruses SARS-CoV and MERS-CoV with high virulence [1-4]. They have pneumonia, ARDS and sepsis [5]. Recent findings shown that, severe COVID-19 patients are strongly associated with a manifestation of lymphopenia, decreased monocytes, eosinophils and basophils, increased neutrophil count [4,6,7]. However, in many cases, hyperimmune reactions were conduct to worst results. For instance, as a result of viral infection the innate immune system of most patients with severe COVID-19 release many pro-inflammatory cytokines and chemokines into circulation. The clinical reflection of this intense reaction so-called ‘cytokine storm’ is seemed as frequently alveolar damage resulting the respiratory failure [4,6-10].
Current approaches on the development of novel therapeutics have been focused on the molecules that regulate of the viral entry. During the viral infection, the S1 subunit of the trimeric S glycoprotein of SARS-CoV-2 recognize its host receptor- ACE2, a type I membrane receptor expressed in various tissues including lungs, heart, kidney and intestine [11,12], while the other subunit, S2, enables fusion of viral and host cell membranes. Most recently, it has been reported that ACE-2-mediated virus internalization is regulated by a serine protease, TMPRSS2 [13]. When SARS-CoV-2 and SARS-CoV were compared in terms of genome and protein similarity, it was revealed that there was similarity between their genome (79%) and protein structure (95-100%). However, it has been determined that the feature specific to SARS-CoV2, but not in other coronaviruses is the furin cleaveage region between the spike protein S1 and S2. Phylogenetic analysis showed that the multibasic amino acid (PRRAR) sequence was included in the relevant segment in the new virus [14] The study of Wrobel et al. shows that the presence of the furin cleavage site in the S protein of SARS-CoV-2 facilitates the conformational change required for the exposure of the RBD domain and binding of the protein to host surface receptors [15].
While functional significance of glycobiology has been well-known for about 40 years, its revolutionary potential on molecular cell biology has been unfolded for last two decades. Emerging advances in glycobiology, and its informatics platforms, glycomics and glycoproteomics, have been enabled it central to development of new strategies for therapeutic and diagnostic purposes [16]. Although different vaccines have been developed against SARS-CoV-2, a more effective treatment strategy has not been developed yet. Glycosylation of surface proteins, one of the virus structural proteins, has an effect on viral entry, impairment of the host's adaptive immunity, and enhancement of infectivity. One of the issues to be clarified is how the proteins of the virus, especially the S protein, are modified by glycosylation mechanisms. In addition, the interaction of the spike protein with ACE2 and furin is extremely important for viral entry. Therefore, revealing the S protein and ACE2 glycosylation profiles and and demonstrating the effect of these glycosylations on virus-host interaction may provide the development of new effective treatment strategies against infection.
In this review, we attempt to summarize current knowledge of mechanism(s) of SARS-CoV-2 infection in terms of glycoperspective. We also mainly focused on the potential glycobiological treatment strategies on the viral infections will be scrutinized.
In the recent studies has been shown that the genome of SARS CoV-2 (NCBI Reference Sequence: NC_045512.2) (Figure 1) is quite similar to the genome of SARS‐CoV-1. (NCBI Reference sequence: NC_004718.3) [17-19]. SARS CoV-2 is an enveloped, positivesense single‐stranded RNA virus and its RNA genome encodes structural and NSPs [20]. SARS-CoV-2 has 11 genes with 11 ORFs. 1. ORF1ab gene expresses 16 NSPs. NSP1 induces host mRNA cleavage. NSP2 binds to host prohibitin 1 and prohibitin 2. NSP3, papain like proteinase, releases NSP1, NSP2 and NSP3 and significant for viral activity. NSP4 which contains transmembrane domain 2 forms an interaction NSP3 and this interaction plays a significant role in viral replication. NSP5, 3C-like proteinase, cleaves at 11 sites of NSP polyprotein. NSP6, putative transmembrane domain, participates in autophagosome formation. NSP7 plays a role in the interaction of NSP8 and NSP12. NSP8 makes heterodimer with NSP7, then NSP12 joins the heterodimer. Thus, RNA polymerase complex is formed. NSP9 binds to helicase cellular protein and this binding is necessary for viral replication. NSP10 may stimulate NSP16, a 2’-O-methyl-transferase. The activity of NSP11 is not yet known. NSP12, RNA dependent RNA polymerase (RdRp), copies viral RNA. NSP13 has helicase activity. NSP14 has 3′ to 5′ endonuclease and N/-methyltransferase activities. NSP15 is endoribonuclease and provides escape from the host immune system. NSP16, 2′‑O‑Ribose‑Methyltransferase, methylates Adenine. Thus, mRNA is protected from host immune system. 2. ORF2 gene expresses S protein which is a glycoprotein and one of the structural proteins of the virus. S protein provides that SARS-CoV-2 attach to host cell membrane receptor. 3. ORF3a gene expresses an ion channel protein related to NLRP3 inflammasome activation. The interaction of this protein with other related proteins results in caspase 1 and IL-1β maturation. 4. ORF4 gene encodes the E protein of the virus. E protein is one of the structural proteins and has the multiple viral functions (e. g. viral assembly, viral replication, viral pathogenesis). 5. ORF5 gene expresses M protein which is structural protein. The role of M protein is viral assembly. 6. ORF6 gene encodes an accessory protein of virus. It has been shown that this protein has the function of viral pathogenesis. 7. ORF7a gene expresses a type I transmembrane protein which is accessory protein. 8. ORF7b gene encodes accessory protein and is expressed in virus-infected cells. 9. ORF8 gene expresses ORF8 protein. 10. ORF9, a structural gene, encodes nucleocapsid (N) protein. 11. ORF10 gene’s function is unknown. Of NSP proteins of SARS-CoV-2, NSP3, NSP5, NSP12 and NSP15 have been suggested to be antiviral drug targets [21].
The binding of viral S proteins to ACE-2 is the first step of coronavirus infection [23]. SARS-CoV-2 S protein (Figure 2) consists of two subunits (S1 and S2) containing SP, N- NTD, RBD, FP, IFP, HR1/2, and TM domains [24]. The proteolytic activation of S protein is essential for viral entry. Cleavage of S protein at the extremely cleavable site that is located at S1/S2 boundary is mediated by furin and is required for binding of RBD of S1 to ACE-2 [25]. In addition to binding, activation of the viral S protein by the host TMPRSS2 is required for virus-cell membrane fusion [26]. The S2 subunit consists of FP, HR1 and HR2 domains. The S2 subunit folds in on itself, bringing HR1 and HR2 domain together. This leads to membrane fusion and then the viral nucleocapsid delivers into the cytoplasm of host cell [23]. The viral RNA is replicated and ORFs are translated to produce viral proteins. Virion assembly occurs in the (ERGIC) (Figure 3). In order to proper virion assembly, viral proteins should undergo a series organized modifications. PTMs of proteins are vital mechanisms for a cell. The main role of PTMs is to functionally modulate proteins with various covalent modifications including phosphorylation, ubiquitination, methylation, acetylation and glycosylation [27]. It is widely confirmed that modification of many viral proteins is depend on host’s PTM machinery [28,29]. The most prominent PTMs of coronaviruses are glycosylation, palmitoylation and phosphorylation of viral proteins [29,30]. Since most of surface proteins of SARS-CoV-2 have been glycosylated, and glycans regulate many cellular processes, glycosylation machinery is essential for the viral functions [31,32].
It has been found in previous studies that coronavirus S proteins covered with N glycosylated motifs [33] that are probable associated with folding of S proteins appositely, priming by host cell proteases as well as modulation of antibody recognition [34]. Together with S1 and S2 subunits, the spike protein of SARS-CoV-2 has 22 [34] known N-linked glycosylation sites, 20 of them are also found in S protein of SARS-CoV-1 [35,36]. At the same time, some glycans in S1 and S2 subunits show homology between SARS-C OV-1 and -2. Receptor binding domain (RBD) of S1 subunit, which is located between residues 303 and 537 [30], is prominent variation site in coronavirus genome [37,38]. More recently two conflicting reports have been published on the structural analysis of RBD. It has been revealed that the most substantial amino acids of RBD for ACE-2 binding in SARS-CoV-2 are L455, F486, Q493, S494, N501 and Y505. However, only one of these residues, Y505, is similar which corresponding region in SARS-CoV-1 [39]. Nevertheless, Zhao’s group declared that the number of amino acids in SARS-CoV-1 RGB that are interfere the binding to ACE-2 is 16 and 8 of them are conserved in SARS-CoV-2 [40]. Although the strong structural similarity between SARSCoV-1 and 2 (approx. 74%) has been revealed, it has most recently been shown that SARS-CoV-2 RBD has larger surface area than 1 and each glycosylation site of S protein in SARS-CoV-2 comprise 22 N-glycans [41] and trace levels of O-linked glycosylation at Thr323/Ser325 (T323/S325) [42] (Table 1). Among these glycosylation sites, RBD comprises only two [40] or three [30,43,44] of them. The revealing of SARS-CoV-2 glycan profiles is extremely important in terms of vaccine and treatment strategies.
N-glycan microheterogeneity, the diversity of N-glycans localizing glycosylation sites of a protein, is essential for orchestrating the protein function. Given the trimeric structure of S protein, it has been conceivable that much site specific N-glycan microheterogeneity may influence function of the S protein which is particularly substantial for anti-viral vaccine development trials [42]. N-linked glycan motifs interfere many cellular events such as inflammatory responses, as well as protein folding and transport [45]. For instance, a phenylalanine residue before the Asparagine-XXX-Serine/Threonine sequon can interacts with first GlcNAc of the N-linked glycan during the glycosylation in ER and the Golgi and eventually can change site-specific N-glycan profile of the protein and also increase the glycosylation efficiency [46-48]. The glycosylation pattern of viral proteins highly depends on glycosylation processes of the host [49]. In SARS-CoV-2 and other human coronaviruses, but not in SARS-CoV-1- related ones, the junctional area of S1 and S2 holds an arginine-rich region that is cleaved at R685-S686 by the cellular protease furin [50] (Figure 2). This polybasic region includes three predicted O-glycosylation sites, S673, T678 and S686, flanking the S1/S2 cleavage region of the S protein. Although the polybasic region would be essential for cleavage of S protein [50], the functional enigma of the predicted O-glycans has not yet been unraveled. Although, the polybasic cleavage site in SARS-CoV-2 is absent in SARS-CoV-1 and other related bat viruses, protease-dependent cleavage is essential for proteolytic maturation of class I viral fusion proteins, that is contribute the fusion potential of virus [51]. More interestingly, O-glycans are quite close the polybasic site in SARS-CoV-2, even the 3rd O-glycan-harbored S (686) is flanked the cleavage site (R687). However, only S336 has predicted O-glycosylation site in S1 of SARS-CoV-1. These results suggest that O-glycan motifs of polybasic cleave site create a masking effect through covering the epitopes of spike protein as previously reported for HIV-1 [52], Ebola [53], Epstein-barr [54], hepatitis C [55], arenavirus [56] and human coronavirus NL63 [57] glycoprotein epitopes. It seems that the predicted O-glycosylation sites that extremely flanked the polybasic cleavage region in S protein may new target for anti-SARS-CoV-2 glycotheraphy.
It should not be surprised that the some of glycoforms that occupy the glycosylation sites in the glycomolecules of host cells, such as furin, ACE-2 and serine protease TMPRSS2, can responsible for virus-host interactions, viral invasion, or activation of envelope proteins such as spike of SARS-CoV-2. Although, in a previous study, Zhou and his colleagues has shown that impairment of ACE- 2 glycosylation machinery can induce to blocking of SARS-CoV-1 entry [58], the glycans in ACE2, furin and / or TMPRSS2 appear to play an important role in SARS-CoV2 infection.
The extracellular domain of the ACE2 receptor contains N and O glycalization sites. There are several O-glycosylated regions and seven N-glycosylated regions including N53, N90, N103, N322, N432, N456 and N690 [12,59]. N90 and N322 of these glycosylation sites have been shown to be important in binding ACE2 to the SARS CoV2 spike protein [60].
The family of proprotein convertases, calcium-dependent serine proteases, are convert precursor proteins into their active forms [61]. As a member of this family, furin is found in mainly epithelial cells, fibroblasts and T-cells as well as endothelial cells of oral mucosa, but it shows a decreased expression profile in B lymphocytes [57]. The spike protein of SRAS-CoV2 has cleavage sites recognized by host cell proteases and required for binding to ACE2. The first cleavage is carried out by the host protease furin (Figure 2) from the region between the S1/S2 subunits boundary of the S protein [25]. Compounds that inhibit furin would be a promising site for COVID-19 treatment [62]. A series of furin inhibitors such as the 4-amidinobenzylamide-derived inhibitors [63], 4-(guanidinomethyl) phenylacetyl-Arg-Val-Arg-4-amidinobenzylamide [64] have been submitted previously, however the knowledge on the unwanted side effects of these compounds remain not being clear [65,66]. Glycan motifs of furin have not been evaluated as target structures for anti-viral drug or vaccine developments so far. Human furin harbours 3 predicted N-glycosylation, N378, N440 and N553, and 33 O-glycosylation sites at different positions (https://www.cbs.dtu.dk/services/NetOGlyc/). Due to their structural, biochemical and conformational dynamics, glycan motifs not only on the viral envelope, but also occupying on the host cells might be prominent motifs for drug binding.
Human type-II transmembrane serine protease, TMPRSS2, is colocalized with ACE-2 (Figure 3) in the surface of epithelial cells and the cleavages of ACE-2 by TMPRSS2 augment the viral fusion [13,67,68]. Previous studies indicated that in the surface of type II pneumocytes and alveolar macrophages, TMPRSS2 co-expressed with 2-6 linked sialic acids that are cellular receptors for influenza virus HA and interfere with N-glycosylation machinery by an unknown mechanism [69,70]. In addition, altered doses of TMPRSS2 have different effects on ACE-2. For instance, low dose of TMPRSS2 converts ACE-2 from an ~130 kDa to an ~115 form whereas as the amount of TMPRSS2 increases, the effect becomes larger [67]. This finding may be evidence that the cell surface glycan motifs are one of the major determinants in viral invasion. TMPRSS2, has two putative N-, N213 and 249 [71], and ten Oglycosylation sites (unpublished data which has reached by using NetOGlyc server, https://www.cbs.dtu.dk/services/NetOGlyc/). However, it has not yet been determined whether these glycosylation sites are involved in the SARS-CoV-2 infection.
In conclusion it can be hypothesized that TMPRSS2 can augment entry of SARS-CoV-2 through converting the glycosylation profile of ACE-2. However, this argument needs further research.
Lectins are a class of specialized glycan-binding proteins which are widely distributed in nature. Apart from antibodies and enzymes, these proteins selectively and reversely recognize carbohydrate molecules through their “carbohydrate recognition motifs” and thereby involve various cellular biological events [72]. On the other hand, Gals are a family of animal lectins that recognize to β-galactoside. They bind cell surface and matrix glycans and regulate many cellular processes [73]. In viral biology, galectins and lectins have various effects such as virus-host interactions, increasing or inhibition of infection and occurring of innate and adaptive immune responses [74]. For instance, DC-SIGN, a C-type lectin found on the surface of dendritic cells, can recognize glycosylated S protein of SARS CoV and facilitate the viral infection through S protein-ACE-2 binding. Similarly, the recognition of S protein by host’s mannose-binding lectin can cause an innate immune response against SARS CoV [75].
Due to their multivalency, in pathogen–host interactions, galectins play in promoting or inhibiting viral infection. Gals can bind to glycan motifs on both virus and host cell surface. According to their binding specificity, either viral infection is prevented or triggered by adhesion to target cell. Therefore, revealing the structure of glycans in viral glycoproteins is important to determine the potential pro- or anti- viral effect of galectins bound to these glycans [76]. Recent reports indicate that Gal-1, Gal-3 and Gal-9 are major galectins that regulate the viral infection. Gal-1, Gal-3 and Gal-9 are major galectins that regulate the viral infection have been reported [76]. For instance, Gal-1 facilitates to gp120 (viral glycoprotein) – CD4+T cell binding in HIV-1. So prevention of interaction between gp120 and Gal-1 might be promised new approaches to HIV-1 therapy [77]. Gal-3 is a potent modulator of cell death in many cell types including T cells, mast cells as well as hepatic stellat cells. It has been suggested that Gal-3's further induction of cell death in HIV-infected cells may be due to the glycosylation change at the Gal-3 receptor [78]. Since galectins have different effects during viral infection, depending on their interaction with viral or host cell molecules, their expression levels, and their types, galectins can be prominent targets for antiviral therapy.
The fact that galectins have different effects depending on their interaction with viral or host cell molecules during viral infection, their expression levels and the type of galectin, makes them a new target in therapy as immune modulators. Mostly, CTD of S1 subunit binds to proteins however NTD prefers extracellular sugars. It has been suggested that the interaction between CTD of SARS CoV-2 spike protein and host ACE2 is depend on galectin-like NTD [79]. More recently, a new type of NTD domain of the SARS CoV-2 spike protein that binds gangliosides has been investigated [80]. This observation indicates that ganglioside associated cell surface sialic acids are crucial for viral entry as well as ACE-2. More interestingly, the spike proteins have similar structures with Gal-3 [81]. Therefore, Gal-3 inhibition that can disrupt the binding of SARS CoV-2 NTD to ganglioside on the cell surface might a new approach for the treatment of COVID-19 [79].
There are two potential targets of anti-viral treatment: viral and cellular components. In both cases the ER and Golgi apparatus play essential roles such as glycosylation machinery, modification of nascent proteins by removing and addition of sugars to core oligosaccharides. ER is also a dynamic organelle for protein quality control that is needful for proper folding, sorting and secretion of newly synthesized proteins. In this complicated system, the glycan motifs introduced as “markers” that are recognized by a series of ER membrane bound lectin chaperons and enzymes such as calnexin, calreticulin, glucosidase I and II [82,83].
During the protein synthesis, all growing glycoprotein harbor three glucose residues at the terminals of their N-linked glycans. Two of three terminal glucoses of improperly folded glycoproteins removed by glucosidases and remaining single glucose serve as a label for recognized by ER membrane-associated lectins, calnexin and calreticulin, hereby incompletely folded glycoproteins containing monoglucosylated N-glycans are arrested in the ER until the glucose is removed by a specific glucosydase. A glycosyl transferase decides whether the protein is properly folded. Improperly folded proteins return to ER and a new glucose is added and the cycle, so-called calnexin / calreticulin cycle, repeats until the glycoprotein has properly folded [82].
A group of carbohydrate mimetics where the substitution of the cyclic oxygen by a nitrogen, iminosugars are potent candidate compounds for the development new anti-viral strategies [84], due to their inhibitor effects on the glucosidase I and II, which mediators of N-glycan processing through the entry of glycoproteins to calnexin/calreticulin cycle [82]. The anti-viral properties of these sugar mimetics have been reported in a series of viral infections such as dengue virus, influenza virus, hepatitis C virus and human immunodeficiency virus and zika virus [84]. The impairment of viral envelope glycoprotein folding due to failure of glucose trimming can reduce of virus-host cell contacts [85,86]. The key point in the anti-viral effects of iminosugars is that the all enveloped viruses have extensively glycosylated proteins that are strongly subjected of calnexin/calreticulin cycle for proper folding. Recent studies indicated that inhibition is strongly based on the iminosugar structure. For instance novel Deoxynojirimycin neoglyco conjugates containing alkyl chains have shown efficiently inhibited a series of glucosidases [85,87]. However, during the chronic viral infection, glucosidase inhibition might lead T-cell activation regarding to the increasing of presentation of peptide epitopes of virus, if given glucosidase inhibitor retains MHC class I in the ER [88].
The answer of this question has not been cleared so far. Previous reports indicated that hemagglutinin antigen of influenza virus is binds sialic acid harboring cell surface molecules with highly affinity and this contact leads viral transduction [89]. Similarly, MERS-CoV have also considered that S glycoprotein shows binding function to cellular sialic acids [90]. These findings can be assumed as sialic acids can mediate SARS-CoV-2 infection [91]. Similar to sialic acid modifications, removing of sialic acids by neuraminidases and mutations in S protein can block viral attachment [91] and hence this can be a potent diagnostic marker for detection of SARS-CoV-2 [92].
The glycan repertoires harbored in most of the viral and cellular glycoproteins serve as signals for the viral invasion processes, such as fusion and endocytosis. In these processes, glycans contribute to viral invasion by a series strategies including shedding of viral glycoproteins from infected cells as snares for the host immune system, escaping from host immune system using with the mimicked glycans (glycomimetics), binding to glycan-recognizing motifs of the host cell or binding to glycan motifs of the host cell via glycan binding receptors such as viral lectins [93,94]. These multi-faced attempts of viruses may explain why we should highlight the glycans in the development of anti-viral strategies. The most popular example of glycan-based drugs are engineered glycan moieties, influenza therapeutics zanamivir (Relenza) [95] and oseltamivir (Tamiflu) [96] which are inhibitor of neuraminidase activity, an enzyme that is essential for virus replication. Although the significance of glycans, synthetic glycans and glycomimetics have recognized, a glyco-strategy for SARS-CoV-2 has still not yet been comprehensively investigated. However, it would appear that engineering glycans will most prominent candidate for the clinical trials to viruses as well as engineered cell surface proteins. However, the main issue is that most of the glycomimetics cannot compete with their natural ligands with sufficient avidity. Consequently multivalent alternative compounds such as polymers and nanoparticles might solve this paucity [97].
No conflict of interest or common interest has been declared by the authors.
 |
Figure 1: The structure of SARS-CoV2 genome [22] |
 |
Figure 2: The structure and furin and S2�cleavage sites of SARS-CoV2 spike protein |
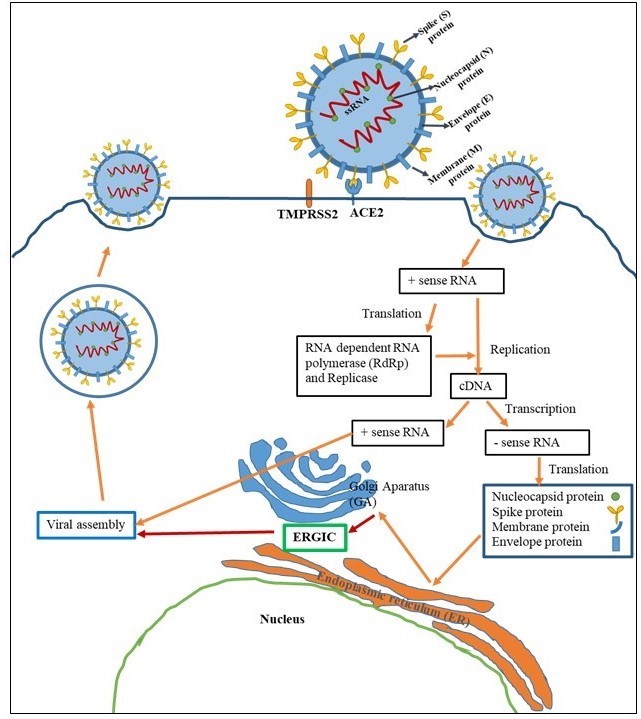 |
Figure 3: Life cycle of SARS-CoV2 |
Glycolysation |
S Protein Domain |
Residue |
Glycan type |
N-linked glycolysation |
NTD |
N17 |
complex |
N61 |
oligomannose- and complex mixture |
||
N74 |
complex |
||
N122 |
oligomannose- and complex mixture |
||
N149 |
complex |
||
N165 |
complex |
||
N234 |
oligomannose |
||
N282 |
complex |
||
RBD |
N331 |
complex |
|
N343 |
complex |
||
RBD to FP |
N603 |
oligomannose- and complex mixture |
|
N616 |
complex |
||
N657 |
complex |
||
N709 |
oligomannose |
||
N717 |
oligomannose- and complex mixture |
||
N801 |
oligomannose- and complex mixture |
||
CH to CD |
N1074 |
oligomannose- and complex mixture |
|
CD |
N1098 |
complex |
|
N1134 |
complex |
||
CD to TM |
N1158 |
complex |
|
N1173 |
complex |
||
N1194 |
complex |
||
O-linked glycolysation |
RBD |
T323 |
|
RBD |
S325 |
|






































